ABOUT US
Your partner in preclinical studies
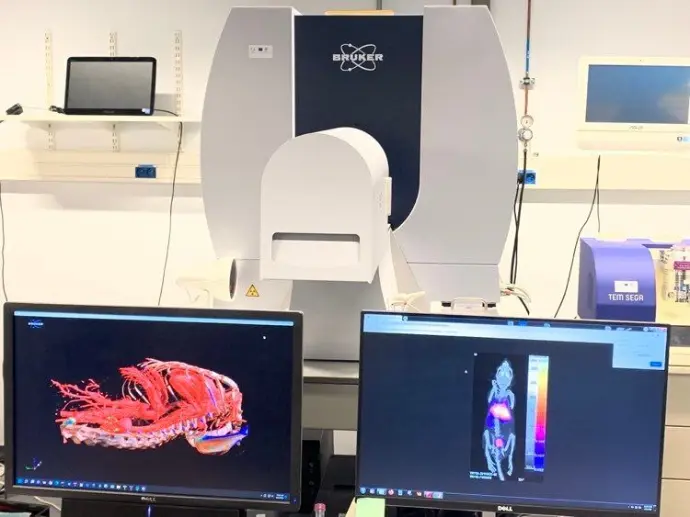
The CMMI is an integrated preclinical imaging facility providing services and training to academic and industrial partners. Our multidisciplinary team offers a wide range of services covering the entire workflow: sample preparation, image acquisition and analysis.
Created jointly by ULB and UMONS with the financial support of the European Union and the Walloon Region (ERDF program), the CMMI is located in the Biopark Charleroi Brussels South, a site that hosts two academic institutes and several companies active in the field of life sciences.
The CMMI will soon join the WalBioImaging network of imaging facilities, an initiative led by ULB, UMONS, CER Groupe, and ULiège, with the support of the Walloon Region and ERDF.


MARTIN Maud, Pr
Scientific co-director ULB

LAURENT Sophie, Pr
Scientific co-director UMONS

MULLER Robert, Pr
Director of international partnerships

SALINITRO Christina
Administrative support

HESS Estelle, PhD
Coordinator
Cutting-edge technology at the heart of Charleroi Biopark
We offer a wide range of preclinical imaging services